Amino Acids are the Building Blocks of Proteins probably lurks right behind Mitochondria is the Powerhouse of the Cell in your subconscious. As a topic on the MCAT, amino acids are incredibly high yield. If you think back to your introductory biochemistry class, you probably remember some of their structures. If you wrack your brain a bit more, you probably remember that amino acids can be classified based on their different properties. Due to how foundational proteins and amino acids are as a topic, knowledge of them is essential for MCAT success. You’re in luck. This article will cut away the fat and teach you how to maximize your performance when dealing with pesky protein questions on the MCAT. At the end, we’ll break down how to read research passages pertaining to the topic as they will appear on the MCAT. Plus, premier instructor Phil Hawkins has created a video masterclass to accompany the article.
|
Before You Dive In: 6 Key Points 1. Fundamental amino acid structure: a basic amino group, an acidic carbonyl group, and a variable “R” group which gives an amino acid its properties. 2. Each amino acids’ structure, name, 1 letter code, 3 letter abbreviation, and class should be memorized. 3. Several amino acids have special properties which appear in specific types of questions. 4. Protein structure can be divided intro primary, secondary, tertiary, and quaternary structure. 5. Protein folding at the level of tertiary structure is affected by many types of interactions and is driven by changes that maximize entropy. 6. Western blotting, SDS-PAGE, and isoelectric focusing are key laboratory techniques used to analyze protein structure and function. PLUS: Watch the overview video below created by JW MCAT instructor Phil Hawkins. |
| Elevate your MCAT prep with an expert tutor
Want expert instruction and guidance for your MCAT prep? Check out our Complete MCAT course or meet with one of our expert MCAT tutors. With tutoring, you can expect a personalized study plan and engaging one-on-one instruction by one of our hand-picked veteran tutors. Check out our courses here or contact us here to get matched with your tutor. |
Description of Amino Acid Structure
Absolute configuration at the α carbon
There are 20 amino acids encoded by the standard human genetic code. 10 of the amino acids are considered essential amino acids for humans since the human body cannot produce them; they must be obtained from the diet. However, knowledge of which amino acids are essential is beyond the scope of what you need to know. Let’s rip the bandage off: you will need to know each amino acids’ name, side chain structure (highlighted in blue below), three letter abbreviation, and one letter abbreviation.

Each amino acid has the same fundamental structure. The central alpha (α) carbon is the glue which holds an amino acid together. The alpha carbon of a free amino acid is bonded to four groups: an amino group (-NH2), a carboxyl group (-COOH), a hydrogen atom, and a unique side chain (-R). Hopefully you’ve taken organic chemistry, because alarm bells should be ringing. By the definition of chirality, an alpha carbon bonded to four unique substituents must be a chiral carbon. In fact, all amino acids are chiral with the exception of glycine, whose R-group is a second hydrogen atom, violating the four unique substituent rule.
The varying side chain (its “R group“) of an amino acid is its money maker. It’s what gives each amino acid its unique properties, influencing its size, polarity, charge, and how it interacts with other parts of a biological system. More on this in the “Classifications” section.

For the 19 amino acids which are chiral, there exist D and L enantiomers which are structurally identical molecules that have opposite absolute configurations around the chiral carbon. As an evolutionary quirk, the human body only uses L-amino acids to synthesize proteins. L-amino acids are those whose amino group points to the left in a Fischer projection (see below). All 19 L-amino acids have an absolute configuration of S, with the exception of cysteine. The alpha carbon of cysteine has an absolute configuration of R.

Amino acids as dipolar ions
The amino group (the “N-terminus“) of an amino acid is a base which is typically protonated in the environment of the cell (which has a pH of 7.4), giving it a positive charge. The carboxylic acid functional group (the “C-terminus“) of an amino acid is an acid which is typically deprotonated in the aqueous environment of the cell, giving it a negative charge. Since both the amino group and the carboxyl group are ionized under physiological conditions, they are by convention depicted as -NH3+ and -COO–. Only the acidic and basic amino acids feature any other ionizable residues. Because the uncharged (non-acidic/basic) amino acids have a plus one and minus one charge under physiological conditions, they are zwitterions (ions whose charges cancel out, producing a molecule with net neutral charge).

Classifications
Like attracts like is more than just dating wisdom. In a biological environment, hydrophilic and hydrophobic interactions are incredibly important. Amino acids can be broadly classified into one of several categories based on their unique R-group, which has important consequences for how they function. Amino acids can be classified as belonging to one of five groups: non-polar aliphatic, non-polar aromatic, polar uncharged, polar basic, and polar acidic.
Non-polar aliphatic amino acids are those whose R-groups contain non-aromatic hydrocarbon chains. Non-polar aromatic amino acids are those whose R-groups contain aromatic rings – except for histidine, which contains an aromatic ring but which is actually classified as a basic amino acid. The non-polar side group of these amino acids means that these amino acids typically interact with other non-polar molecules. For example, non-polar amino acids typically make up the interior of proteins in order to minimize the area with which they are in contact with water. Another example is the plasma membrane of cells: non-polar amino acids typically comprise the hydrophobic interior of a cell membrane, instead of the hydrophilic exterior.
Polar amino acids can be either charged or uncharged. As the opposite of non-polar amino acids, the polar side groups are very hydrophilic and want to maximize their interactions with other polar molecules such as water. Furthermore, they contain atoms capable of hydrogen bonding which further facilitate interactions with water. In a cell, they typically make up the outside of proteins and are often located either in the cytosol or bound to the exterior of membranes. Two of the polar uncharged residues, serine and threonine, have special properties which are listed below.
Polar amino acids can also have a positive or negative charge. Amino acids with acidic residues (aspartic and glutamic acid) have an additional negative charge (aside from the one at their C-terminus) at physiological pH, while amino acids with basic residues (lysine and arginine) have an additional positive charge (aside from the one at their N-terminus) at physiological pH. Note that histidine is an exception. Because it contains a basic residue that is deprotonated under physiological conditions, histidine is neutral at physiological pH. The pKa values of the acidic and basic residues, as well as of the N and C termini, are shown below.

Amino acids are not always kept under physiological conditions, where pH is about 7.4. Changes in the pH of their environment can affect their charge. When amino acids are kept under low pH conditions, more of the residues will be protonated, resulting in a net positive charge. When amino acids are kept under high pH conditions, more of the residues will be deprotonated, resulting in a net negative charge.
Special Properties of Side Chains
We will quickly overview some of the most important cases where an amino acid side chain gives that amino acid a unique property; memorize each side chain and be ready to apply their properties on test day.
1. Some amino acid side chains naturally destabilize proteins with a helical structure. Specifically, if glycine or proline are added into a protein, they will often destabilize (break down) local alpha helical secondary structure.
2. There are several amino acids whose side groups are most often targets of phosphorylation, including serine, threonine, and tyrosine. Phosphorylation is the addition of a negatively charged phosphate to a molecule, which often changes the structure or function of that molecule. Notice that each of these three amino acids features a hydroxyl functional group. Histidine is often a target of phosphorylation, but this information does not seem to be considered testable by the AAMC.

3. Several amino acids can mimic a permanently phosphorylated functional group. Aspartic acid and glutamic acid have a structure that is similar enough to phosphate that, when substituted for serine/threonine/tyrosine, are able to mimic the presence of a phosphate group. When an enzyme is activated by phosphorylation, mutating in an aspartic acid residue could therefore cause it to become permanently activated. This is known as the phosphomimetic effect.

4. Cysteine is sometimes able to produce covalent disulfide bonds with other cysteine residues, holding together the subunits of a polypeptide.
5. Basic and acidic residues can interact to form salt bridges due to their attractive positive and negative charges. Salt bridges are an attractive interaction which is a combination of hydrogen bonding and acid/base interaction; they play an important role in stabilizing proteins.
Synthesis of α-amino acids
There are two primary methods for synthesizing amino acids in the laboratory and amino acid precursors with which you should vaguely understand: the Strecker synthesis and the Gabriel synthesis. Definitely don’t memorize intimate details, but be familiar with the overall idea and some key takeaways.
Strecker Synthesis
The Strecker Synthesis is a two-step pathway in which different types of amino acids can be synthesized. In the first step, ammonia (NH3) is added to an aldehyde (with a variable R group – this is important later), forming an imine. Next, cyanide ion nucleophilically attacks the imine, forming an alpha-aminonitrile. In the second step, the aminonitrile is hydrolyzed, resulting in an alpha-amino acid. By varying the R group on the aldehyde, we can synthesize a variety of alpha amino acids with their own particular R-groups. Importantly, since cyanide ion is added to the imine non-stereospecifically, the Strecker synthesis produces a racemic mixture of L and D amino acids (in other words, 50% L and 50% D). Researchers will need to use an enantioselective method for isolating just the L or D enantiomer.

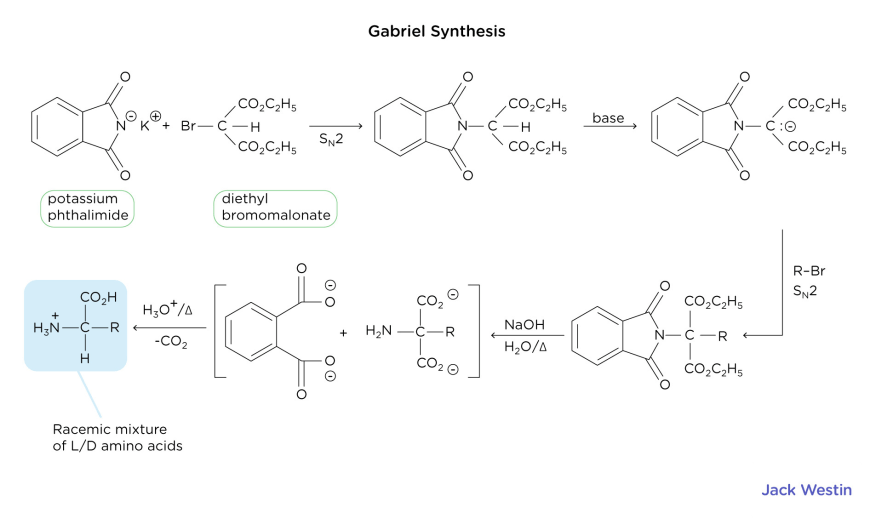
Gabriel Synthesis
The Gabriel Synthesis should be known in a little bit more depth due to the intricacies of its synthesis. The nitrogen of a phthalimide salt is modified with a diethyl bromomalonate. The phthalimide substituted malonic ester (the first product shown) has an acidic hydrogen (due to stabilization of the anion by the two nearby ester groups). This hydrogen can be deprotonated, and the resultant anion can be alkylated by a molecule with a variable R-group (which is what allows for the creation of different amino acids). The phthalamide undergoes base catalyzed hydrolysis; the product is decarboxylated with heat and acid to form a chiral amino acid. In this case, since the decarboxylation reaction is non-stereospecific but produces a chiral amino acid, the product of decarboxylation is also a racemic mixture of L and D amino acids. In other words, both the Strecker and Gabriel synthesis reactions result in a racemic mixture.
Amino Acids Practice Quiz
Now that you’ve read up on all of the amino acids and they’re properties, stop and take this Amino Acid Practice Quiz to test your knowledge.
Reactions
Sulfur linkage for cysteine and cystine
A covalent disulfide bond (sulfur linkage) can form between the sulfur-containing R groups of two cysteine molecules (called sulfhydryl groups). Disulfide bonds between cysteine residues can affect protein folding and stability. Disulfide bonds form between cysteine residues under oxidizing (high pH) conditions. Disulfide bonds can be broken under reducing (low pH) conditions. In the laboratory, researchers often use beta mercaptoethanol (BME) to break down the disulfide bonds of a protein during a western blot.

Peptide linkage: polypeptides and proteins
A peptide is a molecule composed of two or more amino acids. The bond connecting together the two amino acids is a peptide bond. It occurs when the amino group of one amino acid nucleophilically attacks the carboxyl group carbon of another amino acid, linking the two molecules together and releasing a water molecule. In the final product, the amino and carboxyl group have been transformed into an amide functional group (carbon double bonded to an oxygen and single bonded to a nitrogen). Because this reaction releases a water molecule, it is a specific example of a condensation reaction. Peptide bond formation is endergonic (a process which requires energy), the energy for which typically comes from ATP.

Hydrolysis
Long chain polypeptides can be formed by linking many amino acids to each other via peptide bonds. The peptide bond can only be broken by hydrolysis, where the bonds are cleaved with the addition of a water molecule. Because this reaction is the reverse of peptide bond formation, it is exergonic (releases energy) and occurs spontaneously. Despite the fact that peptide bonds will spontaneously want to break down, the activation energy for this reaction is high enough that peptide bonds are metastable and will break down very slowly. Living organisms have enzymes that are capable of both forming and breaking peptide bonds.
General Principles of Protein Structure
Now that we’ve covered the fundamentals of amino acids, let’s lay the groundwork for understanding their higher order counterpart: proteins. For you physicians who aspire to be bodybuilders (cough, orthopedic surgeons), this is the good stuff.
Primary structure of proteins
Amino acids can be connected end to end from N-terminus to C-terminus, forming longer chains of amino acids. The linear connectivity of amino acids is the primary structure of a protein. The primary structure of a protein can essentially be thought of as the sequence of amino acids starting at an N-terminus (the amino group of the very first amino acid in the chain) and ending at a C-terminus (the free carboxylic acid group of the very last amino acid in the sequence). In a single peptide, each amino acid is specified with a number. The amino acid at the N-terminus is numbered 1, the one adjacent is numbered 2, and so on until the C-terminus is reached.

Secondary structure of proteins
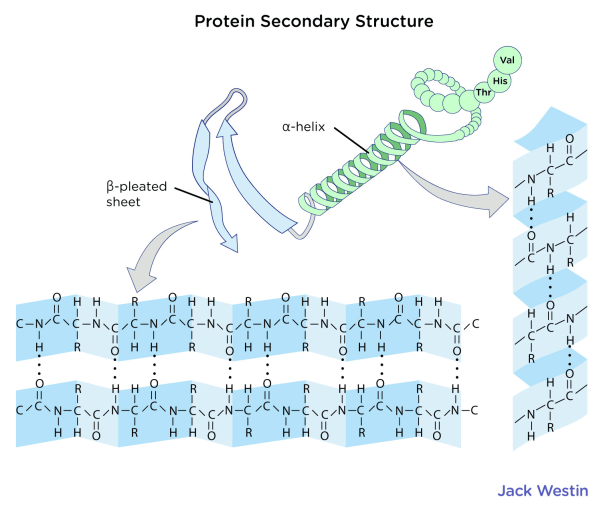
Once a single chain of amino acids is produced, that chain of amino acids will fold (like a string folding into a ball of yarn) in a way that maximizes the stability of the protein and maximizes the entropy of the system. We’ll dive deeper into protein folding later on; for now this description is sufficient. In a very long chain of amino acids, smaller regions will locally fold into special structures, most often either beta sheets or alpha helices.
Alpha helices are helical structures which have a variety of roles such as acting as DNA binding sequences or anchoring larger proteins (such as G-protein coupled receptors) to the plasma membrane by acting as a transmembrane sequence (a region of a protein which passes through the plasma membrane). The alpha helix is stabilized by hydrogen bonding interactions between peptide backbone carbonyl (C=O) oxygen atoms and peptide backbone amide groups 4 residues away (i, i+4). Hydrogen bonding is the primary stabilizing force in protein secondary structure.
Beta sheets are sheetlike structures created by several regions of a peptide chain looping back along one another; these regions interact through hydrogen bonding interactions between backbone carbonyl and amino groups. The regions where the peptide chain loops back on itself are called beta turns.
Tertiary structure of proteins
The overall three dimensional structure that a peptide chain folds into is its tertiary structure. Once all the secondary structure elements of a peptide have folded, the peptide will fold into the most energetically favorable state that it can reach; this most optimal conformation is called a peptide’s native state. When protein folding takes place in the aqueous environment of the body, the hydrophobic R groups of nonpolar amino acids mostly lie in the interior of the protein (to minimize their contact with water), while the hydrophilic R groups lie mostly on the outside (to maximize their contact with water). Internally, the peptide is stabilized by hydrogen bonding, ionic bonding/salt bridges, dipole-dipole interactions, London dispersion forces, and disulfide linkages. Hydrophobic interactions occur between hydrophobic residues (such as the nonpolar aliphatic and aromatic side chains) as they try to maximize their contact with one another and minimize their contact with water. Disulfide linkages between cysteines are the only covalent bond capable of influencing tertiary structure (as the covalent peptide bonds have already formed) All interactions, weak and strong, determine the final three-dimensional shape of the protein. When a protein loses its three-dimensional shape, its function will be lost or altered unless it refolds.

Quaternary structure of proteins
A single chain of amino acids will fold into a single peptide; the combination of multiple peptides together, or the combination of peptides with non-protein molecules, is the quaternary structure of a protein. When multiple peptide subunits join together, they form a polypeptide. There are special terms for describing the arrangement of multiple peptides together. A single peptide alone is termed a monomer, two peptides joined together are termed a dimer, three peptides joined together are termed a trimer and so on. If the peptides joined together are identical, they receive the prefix “homo-“, while if the peptides are dissimilar they receive the prefix “hetero-“. For instance, two identical peptides joined together are a homodimer, while three different peptides joined together are a heterotrimer.
Peptides can also be joined to non-protein molecules called prosthetic groups. Prosthetic groups are non-protein molecules which can be organic (i.e. carbs, lipids, vitamins) or inorganic (e.g. metal ions) that are tightly bound to a peptide and are required for protein functionality. An apoprotein or apoenzyme is a protein without its characteristic prosthetic group; apoproteins are generally nonfunctional. An apoprotein that has been joined with its prosthetic group and gained functionality is a holoprotein.

Conformational stability
Why do proteins fold into the shapes that they do? Furthermore, what determines whether they stay that way?
Denaturing and folding
The stability of a protein depends on its environment, its intermolecular interactions, and its intramolecular interactions. Proteins fold into a state that maximizes the entropy of the system and minimizes their own intrinsic energy. A low energy protein is a more stable protein. A protein will spontaneously fold into lower and lower energy conformations. On its way, a protein may transiently form a molten globule, which is a protein that has a relatively folded tertiary structure but lacks the functionality of a native state protein. In some cases, a protein may need extra assistance from chaperone proteins which assist in folding parts of a protein. If their is a high energetic barrier for escaping from the molten globule state, a chaperone protein may be used to get over the energetic hump, so to speak.
The exact, most stable structure of a protein depends on the environment of that protein. Many factors in the environment, such as salinity, temperature, and pH, will affect the folding of a protein. In an aqueous environment, hydrophilic residues will want to be positioned on the outside of a protein. At a high pH, more of the proteins residues will be deprotonated. At high salinity, internal ionic interactions within the protein will be disrupted, impeding folding. A protein will fold into its native state when the conditions are optimal for it to do so, and changing any of these factors may change the final tertiary structure into which a protein folds.

When a protein is folded into its native state, changing elements of the environment can cause it to denature, or unfold. Changing the temperature, salinity, or pH away from their optimal values will result in unfolding of a native state protein into a denatured and functionally inactive form. It is often possible to reverse denaturation because the primary structure of the polypeptide (the covalent bonds holding the chain of amino acids in their correct sequence) is intact. Once a factor causing denaturing is removed, the protein will usually spontaneously refold into its native state.

Hydrophobic interactions and the solvation layer (entropy)
Proteins largely function in an aqueous environment. Therefore, hydrophobic interactions are important in imparting stability to a protein. During protein folding, amino acids with nonpolar, hydrophobic R groups are sequestered on the inside of a protein, away from water. On the interior of the protein, hydrophobic R groups are able to interact favorably to maximize the stability of the protein.
Entropy is the extent to which a system is disordered, spread out, and chaotic. To make a long story short, the universe wants more entropy. Because a folded protein is more ordered, a folded protein has less entropy than an unfolded protein. If this is the case, then why is protein folding entropically favorable overall? It’s because a protein does not fold in a vacuum. The decrease in entropy due to protein folding is balanced out by an increase in the entropy of the surrounding water molecules who no longer have to interact with the hydrophobic residues on the interior. The water molecules surrounding the protein (a protein’s solvation layer) are able to have fewer unfavorable interactions with hydrophobic residues and more favorable interactions with hydrophilic amino acids, maximizing the total entropy of the system.

Separation techniques
Isoelectric focusing
The isoelectric point (pI) of a protein is the pH at which the net charge on a protein equals zero. At a pH above the proteins pI (a relatively alkaline pH), a protein will be negatively charged, while at a pH below the proteins pI (a relatively acidic pH) a protein will be positively charged. The presence of additional acidic groups (such as the side chains of aspartic and glutamic acid), which have low pKa values, will lower a protein’s pI. The presence of additional basic groups (such as the side chains of lysine and arginine), which have high pKa values, will increase a protein’s pI.

The isoelectric point of a protein can be determined using isoelectric focusing. In this technique, a pH gradient is created on top of a membrane. One end of the membrane is connected to a negatively charged cathode, while the other end of the membrane is connected to a positively charged anode. When proteins are dolloped onto the membrane, their baseline charge will determine whether they will be attracted to the cathode or to the anode. Each protein will migrate until it reaches the location along the pH gradient which is equal to that protein’s isoelectric point, at which point the protein will be neutral and will no longer be attracted to either the cathode or anode. The protein will remain stationary once it has reached its isoelectric point; visualization of the protein’s location will provide researcher’s with information about the pI of that protein.

Electrophoresis and Western Blot
Native PAGE or SDS-PAGE are used in conjunction with Western Blotting to analyze the structure of proteins or the expression of proteins in a cell. Native polyacrylamide gel electrophoresis (Native PAGE) is a laboratory technique which separates proteins in their native tertiary-state on the basis of size and charge. Proteins are placed on a porous polyacrylamide gel membrane and loaded near the negatively charged cathode. The proteins then migrate toward the positively charged anode at the opposite end of the gel. Smaller molecules move through the pores in the gel faster than larger molecules, and this difference in the rate of migration separates the fragments based on size. There are molecular weight standard samples that can be run alongside the molecules to provide a size comparison.
SDS-PAGE is a variation of the PAGE electrophoresis method which separates proteins only by molecular mass, not charge. Sodium dodecyl sulfate (SDS) is an amphipathic detergent which when applied to protein binds throughout all parts of the protein, causing the protein to completely denature and unfold from its tertiary structure into a straight chain. The denatured protein obtains a uniform mass to charge ratio which allows the protein to be separated only on the basis of its mass, instead of both its mass and charge. Optionally, researchers can run an SDS-PAGE gel under reducing conditions by adding beta-mercaptoethanol (BME), a reducing agent which breaks down disulfide bonds in a protein. Since SDS is able to break down all non-covalent bonds within a protein and BME is able to break down all intermolecular and intramolecular disulfide bonds, SDS-PAGE under reducing conditions can be used to separate a completely denatured and isolated monomeric subunit of a peptide.

Once proteins have been separated using Native or SDS PAGE, they can be visualized using Western Blot. During a Western Blot, proteins are transferred from the polyacrylamide membrane to a blotting membrane. Afterwords, highly specific radioactive antibodies can be added to the membrane to visualize the proteins present or detect whether a particular protein is present on the gel.

How to Read MCAT Research Passages on Proteins
The core skill required to succeed on the MCAT is the ability to quickly analyze research passages. Here we’re going to cover how to quickly break apart common figures and diagrams found on the MCAT. The takeaway will be that breaking down research is much easier than it looks and something everyone can do. When amino acid based research passages pop up, they most commonly contain either pictures of amino acids and peptides, which must be visually understood, or graphs and tables describing the effects of amino acid mutations, which must be analyzed.

Figure 1 HIV-1 protease (PR) catalytic triad (Gly/Thr/Asp) active site containing a peptide (25/26/27 and 25’/26’/27’ refer to amino acids of two different HIV-1 PR monomers)
We’ll cover the former using the image above, which is an example of something the MCAT might show you. Notice that the figure has a caption which is essential for understanding the contents of the image. The caption states that this is an “HIV-1 protease catalytic triad (Gly/Thr/Asp active site containing a peptide.” At least two things should be clear based on the caption; one, this is the active site of an enzyme, which is where a substrate binds during a reaction, and two, the amino acids glycine, threonine, and aspartic acid are involved in catalysis. In most images of molecules, solid lines typically represent intramolecular covalent bonds and dotted lines typically represent intermolecular forces. Therefore, we can perhaps infer that the bottom part of the image is one molecule, the middle part of the image is another molecule, and the top part is yet another, each of the three connected by hydrogen bonds. We can also see two water molecules (H2O) which have entered the active site and which are assisting in coordinating everything together. The middle part of the molecule is most likely the substrate, based on our knowledge that this is the active site of the enzyme HIV-1 PR.
Most importantly, let’s notice the labeled amino acids. Whoever created the image thought it was important enough to add the labels “Gly27”, “Thr26”, “Asp25”. Each of these labels refers to the amino acid and the position within the protein that an amino acid is located. Remember that proteins are just long chains of amino acids connected by peptide bonds. The amino acids are numbered starting at 1 for the carbon at the N-terminal side of a protein. Asp25 refers to Aspartic Acid at position 25, Thr26 refers to Threonine at position 26, and so on. Noting the type of amino acid at each position is important because it allows us to make predictions. For example, would it be beneficial or harmful to the enzyme to replace the amino acid at position 25 with arginine? Well, arginine is positively charged, and would have the exact opposite effect as the negatively charged aspartic acid. Therefore it would most likely be harmful. Now, we’ll take a look at the table below.

Table 1 Catalytic activity of wildtype and mutant HIV-1 protease (PR)
The caption states that this is a table showing the catalytic activity of either the wildtype (normal) enzyme or mutant versions of the enzyme. We know that the “variant” column is the independent variable because it’s something that researchers change – they create a mutant version of an enzyme. The “activity” column is the dependent variable because it’s something that researchers measure a change in – how much has enzyme activity changed from baseline? Therefore, the table is answering the question: when a mutation is made within the structure of an enzyme, how is enzyme activity affected? Don’t worry about the units too much unless asked about it, it’s often enough to qualitatively grasp what a table is presenting you and to notice trends in how a dependent variable changes in response to a change in an independent variable.
The normal (wildtype) enzyme has an activity of 35 M/s. The mutant labeled “G27V” has an activity of 33 M/s. Let’s get some naming principles out of the way – G27V is a way of describing what mutation was made. The first letter refers to the amino acid which was originally present, the number refers to the position within the protein, and the last letter refers to the new amino acid. In this case, G27V refers to the glycine (G) at position 27 being replaced with valine (V). Apparently, when this happens the activity doesn’t change much. Therefore, we can conclude that when glycine at position 27 is substituted with valine, enzyme activity doesn’t differ much from normal.
D25E is a mutant enzyme where the aspartic acid at position 25 was substituted with glutamic acid. In this case activity is greater in the mutant, which means that this was a helpful mutation. D25R is a substitution at the same position but this time for arginine. In this case, activity is less in the mutant, which means that this mutation negatively impacted enzyme activity. Therefore, if a question asked what kind of mutation most negatively impacts the activity of HIV-1 PR, the answer would be along the lines of substituting a negatively charged residue (D) for a positively charged residue (R).
Practice Questions for Proteins and Amino Acids
Jack Westin Practice
- Complete the Jack Westin Amino Acid Practice Quiz 1
- Complete the Jack Westin Amino Acid Practice Quiz 2
- Complete the Jack Westin Amino Acid Practice Quiz 3
- Complete the Jack Westin Protein Structure Quiz 1
- Complete the Jack Westin Protein Structure Quiz 2
- Proteins and Fluorescence Practice Passage by Jack Westin
Khan Academy Practice
- Collagen Mutations Result in Osteogenesis Imperfecta
- Glucogenic and ketogenic amino acids
- What causes blood cells to sickle?
AAMC Practice
- Official Guide C/P Section Passage 1 Question 2
- Chemistry Question Pack Question 37
- Online Flashcard Biochemistry 14
- Online Flashcard Biochemistry 22
- Practice Exam 3 B/B Section Passage 5 Question 27
- Chemistry Online Flashcards Question 24
- Section Bank C/P Section Passage 10 Question 74
- Practice Exam 2 B/B Section Passage 2 Question 6
Key Terms
- Amino acid: simple organic structure which is the fundamental building block of proteins and is identifiable based on a unique R group
- Alpha carbon: the carbon adjacent to the carbonyl carbon of an amino acid to which the amino, carboxyl, R, and hydrogen groups are attached
- R group: the unique functional group attached to the alpha carbon of each type of amino acid; confers each amino acid its special properties
- L and D enantiomers: the two enantiomeric forms of amino acids; humans have only L amino acids
- N terminus: the end of an amino acid containing the positively charged amino group
- C terminus: the end of an amino acid containing the negatively charged carboxyl group
- Zwitterion: a molecule that is net neutral because it bears an equal amount of positive and negative charge
- Non-polar aliphatic: the amino acids which have an R group that is primarily a non-polar straight chain hydrocarbon (i.e. made of C and H atoms)
- Non-polar aromatic: the amino acids which have an R group that is an aromatic ring (except for histidine)
- Polar uncharged: the amino acids which have an R group that is polar but neutrally charged
- Acidic: the amino acids which have an R group that is a hydrogen donor and is negatively charged under physiological conditions
- Basic: the amino acids which have an R group that is a hydrogen acceptor and is positively charged under physiological conditions
- Phosphorylation: the addition of a phosphate group (PO43-) to a molecule
- Phosphomimetic effect: the carboxylic acid R group of aspartic and glutamic acid can mimic the presence of a phosphate group
- Strecker Synthesis: synthesis of a racemic mixture of L and D amino acids from an aldehyde precursor
- Gabriel Synthesis: synthesis of a racemic mixture of L and D amino acids from a phthalamide precursor
- Disulfide bond: a covalent bond between the sulfhydryl R groups of two cysteine amino acids
- Salt bridge: an attractive interaction which is a combination of hydrogen bonding and acid/base interaction
- Peptide bond: a covalent amide linkage between the N terminus and C terminus of two amino acids
- Amide: a functional group in which a nitrogen is bonded to a carbonyl carbon atom
- Condensation: a bond forming reaction in which water is lost as a byproduct
- Hydrolysis: a bond breaking reaction which is carried out through the addition of water
- Primary Structure: The sequence in which amino acids are linearly connected
- Secondary Structure: The small alpha helix and beta sheet structures into which a peptide locally folds
- Beta Sheet: A sheetlike element of protein secondary structure
- Alpha Helix: A helical element of protein secondary structure
- Tertiary Structure: The three dimensional shape into which a protein folds overall
- Native State: The tertiary structure of a protein in which it is biologically active
- Quaternary Structure: The connectivity of a peptide with other peptides (forming a polypeptide) or prosthetic groups
- Prosthetic Group: A non-protein molecule which is permanently attached to a protein and is required for protein function
- Apoprotein: A protein that requires a prosthetic group but is lacking that prosthetic group
- Holoprotein: An apoprotein bound to its prosthetic group
- Chaperone proteins: A protein which assists in the folding of other proteins’ tertiary structure
- Denaturation: The unfolding of a protein due to suboptimal environmental conditions
- Solvation layer: The layer of solvent interfacing with the exterior of a protein
- Isoelectric Point: The pH at which a molecule is neutral (also called its pI)
- Isoelectric focusing: A laboratory technique in which a protein migrates along a membrane until it reaches its pI
- Native PAGE: A laboratory technique for separating proteins in their native state tertiary structure by charge and size
- SDS PAGE: A laboratory technique for separating proteins by size alone
- Western Blot: A laboratory technique for visualizing proteins after they have been separated using PAGE

